Using MATLAB Parallel Server
Why Use Matlab Parallel Server
For many Matlab users, they are very familiar with running things from the interactive Matlab Graphical User Interface (GUI). Although you can launch the GUI on Raj, there are several reasons why it is not an ideal workflow, most prominent of which is the lag introduced by running Matlab over X11 forwarding and sometimes even over the VPN. The most effective way to run a Matlab script on Raj is to use a Slurm submission script. However, learning to run Matlab this way can be challenging. That is where Matlab Parallel Server comes in. This toolbox gives users the ability to run jobs on Raj from their home Desktop installation of Matlab by integrating with Slurm on the backend. This allows for near seamless transition from testing scripts on your Desktop to running them on the cluster, without needing to learn all the nuances of writing a Slurm submission script.
Configuring Your Matlab Profile on Raj
After logging into the cluster, you will need to create a Matlab parallel server profile. To do this you need to add some scripts to your default Matlab path. First some background information on how Matlab operates. When you launch Matlab, it sets up a list of directories where it searches for scripts and functions which can be called from the command window. Most of these directories exist within the Matlab installation directory, however, Matlab adds an extra directory for custom scripts from each user inside their home directory. This directory is located at $HOME/Documents/MATLAB. In order to add the files necessary to run Matlab Parallel Server, you need to create this directory (if you have not already done so) and copy the necessary files into it. To do this run the following commands:
mkdir -p $HOME/Documents/MATLAB
tar -xf /cm/shared/Public/matlab-parallel-server.tar.gz -C $HOME/Documents/MATLAB
You should now have a MATLAB directory in your home directory which is populated with the files needed to configure your Matlab parallel profile. Now load a version of Matlab you wish to configure and launch Matlab. Note that you need to configure each version of Matlab you wish to use and the version you run on Raj, must match the version you are using on your Desktop.
module load matlab/R20xxx
matlab
From the matlab command window run the command configCluster. You should get an output which says:
Complete. Default cluster profile set to "raj R20xxx"
Your cluster profile is now set up. Note that this step only needs to be done once per version. Once your profile is configured there should be no need to reconfigure it.
Configuring your Desktop Environment
Configuring your Desktop environment is very similar to configuring your cluster profile on Raj. First you need to download the necessary zip/tarball archive. You can download the files here. Next start Matlab and untar/unzip the directory specified by calling the Matlab command userpath. Now you can configure your system using the Matlab command configCluster. Your system is now set up. Note that by default, the configCluster command defaults all parpool jobs to run remotely on Raj. You run local jobs by specifying the 'local' profile in your parallel matlab commands. Additionally, you can reset your local profile to be the default profile by updating the settings found in the Matlab menu parallel → create and manage clusters.
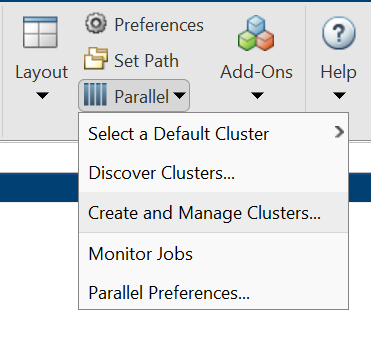
Configuring Jobs (optional)
Prior to submitting the job, we can specify various parameters to pass to our jobs, such as queue, e-mail, walltime, etc. The following is a partial list of parameters. See AdditionalProperties for the complete list.
>> % Get a handle to the cluster
>> c = parcluster;
Now that we have a cluster handle we can specify our parameters.
>> % Specify the account to use
>> c.AdditionalProperties.AccountName = 'account-name';
>> % Request email notification of job status
>> c.AdditionalProperties.EmailAddress = 'user-id@marquette.edu';
>> % Specify number of GPUs to use
>> c.AdditionalProperties.GpusPerNode = 1;
>> % Specify memory to use, per core (default: 4gb)
>> c.AdditionalProperties.MemUsage = '6gb';
>> % Specify the queue to use
>> c.AdditionalProperties.QueueName = 'queue-name';
>> % Specify the wall time (e.g., 5 hours)
>> c.AdditionalProperties.WallTime = '05:00:00';
If you want you can save the changes after modifying AdditionalProperties for the above changes to persist between Matlab sessions.
>> c.saveProfile
To see the values of the current configuration options, display AdditionalProperties.
>> % To view current properties
>> c.AdditionalProperties
Unset a value when no longer needed.
>> % Turn off email notifications
>> c.AdditionalProperties.EmailAddress = '';
>> c.saveProfile
Running Batch Jobs
Use the batch command to submit asynchronous jobs to the cluster. The batch command will return a job object which is used to access the output of the submitted job. See the Matlab documentation for more help on batch.
>> % Get a handle to the cluster
>> c = parcluster;
>> % Submit job to query where MATLAB is running on the cluster
>> job = c.batch(@pwd, 1, {}, ...
'CurrentFolder','.', 'AutoAddClientPath',false);
>> % Query job for state
>> job.State
>> % If state is finished, fetch the results
>> job.fetchOutputs{:}
>> % Delete the job after results are no longer needed
>> job.delete
To retrieve a list of currently running or completed jobs, call parcluster to retrieve the cluster object. The cluster object stores an array of jobs that were run, are running, or are queued to run. This allows us to fetch the results of completed jobs. Retrieve and view the list of jobs as shown below.
>> c = parcluster;
>> jobs = c.Jobs;
Once we’ve identified the job we want, we can retrieve the results as we’ve done previously. fetchOutputs is used to retrieve function output arguments; if calling batch with a script, use load instead. Data that has been written to files on the cluster needs be retrieved directly from the file system (e.g. via ftp).
To view results of a previously completed job:
>> c = parcluster;
>> jobs = c.Jobs;
Note: You can view a list of your jobs, as well as their IDs, using the above c.Jobs command. Also note that this is a Matlab job ID not the Slurm Job ID used on Raj.
>> % Fetch results for job with ID 2
>> job2.fetchOutputs{:}
Parallel Batch Jobs
Users can also submit parallel workflows with the batch command. Let’s use the following example for a parallel job, which is saved as parallel_example.m.
function [t, A] = parallel_example(iter)
if nargin==0
iter = 8;
end
disp('Start sim')
t0 = tic;
parfor idx = 1:iter
A(idx) = idx;
pause(2)
idx
end
t = toc(t0);
disp('Sim completed')
save RESULTS A
end
This time when we use the batch command, to run a parallel job, we’ll also specify a Matlab Pool.
>> % Get a handle to the cluster
>> c = parcluster;
>> % Submit a batch pool job using 4 workers for 16 simulations
>> job = c.batch(@parallel_example, 1, {16}, 'Pool',4, ...
'CurrentFolder','.', 'AutoAddClientPath',false);
>> % View current job status
>> job.State
>> % Fetch the results after a finished state is retrieved
>> job.fetchOutputs{:}
ans =
8.8872
The job ran in 8.89 seconds using four workers. Note that these jobs will always request N+1 CPU cores, since one worker is required to manage the batch job and pool of workers. For example, a job that needs eight workers will consume nine CPU cores.
We’ll run the same simulation but increase the Pool size. This time, to retrieve the results later, we’ll keep track of the job ID.
NOTE: For some applications, there will be a diminishing return when allocating too many workers, as the overhead may exceed computation time.
>> % Get a handle to the cluster
>> c = parcluster;
>> % Submit a batch pool job using 8 workers for 16 simulations
>> job = c.batch(@parallel_example, 1, {16}, 'Pool', 8, ...
'CurrentFolder','.', 'AutoAddClientPath',false);
>> % Get the job ID
>> id = job.ID
id =
4
>> % Clear job from workspace (as though we quit MATLAB)
>> clear job
Once we have a handle to the cluster, we’ll call the findJob method to search for the job with the specified job ID.
>> % Get a handle to the cluster
>> c = parcluster;
>> % Find the old job
>> job = c.findJob('ID', 4);
>> % Retrieve the state of the job
>> job.State
ans =
finished
>> % Fetch the results
>> job.fetchOutputs{:};
ans =
4.7270
The job now runs in 4.73 seconds using eight workers. Run code with different number of workers to determine the ideal number to use.
Alternatively, to retrieve job results via a graphical user interface, use the Job Monitor (Parallel → Monitor Jobs).
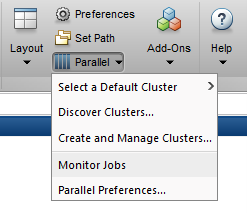
Prestaging Data
By default, Matlab copies all necessary file dependencies up to the cluster. When using functions and small files, this works well. However, say you have a large data set you want to run some example on. Depending on the size of the data set, it could take several minutes to several hours to copy the data to Raj. For this reason, it can be prudent to copy the data up to Raj prior to submitting the job. This process is called prestaging your data. This can be achieved by using the AdditionalPaths attribute of your parcluster object.
Let's follow an example which calculates the eigen values of the prestaged file, x.mat, located at $HOME/MATLAB/data/x.mat. First, let's look at the code used to create x.mat.
>> x = rand(1000, 1000, 150);
>> save('x.mat','-mat', '-v7.3','x')
Now let's look at the example script which is saved as prestage_example.m.
% Load data
load('x.mat');
% pre allocate vars
N = size(x,3);
a = zeros(N,1);
b = zeros(N,1);
% run in series
tic;
for I = 1:N
a(I) = max(eig(x(:,:,I)));
end
t1 = toc;
% run in parallel
tic
parfor I = 1:N
b(I) = max(eig(x(:,:,I)));
end
t2 = toc;
% clear x so it does not get downloaded with fetchOutputs
clear x
To prestage data first we need to create a location on Raj where you will keep your data. In this example we will use $HOME/MATLAB/Data.
cd $HOME/MATLAB
mkdir Data
Next copy your data to the Raj using your preferred method (e.g. sftp, scp, rsync, etc).
Finally, let's run our cluster job.
NOTE: Since we are running a script instead of a function the batch notation is a bit different. Instead of @function we use 'scriptname' (note the lack of the .m extension in both formats), additionally we drop the arguments specifying number of outputs, and the curly braced set of input arguments. In the previous example those were specified as 1 and {16}.
>> % start cluster
>> c = parcluster;
>> % define path to x.mat
>> % Note: change <username> to your Raj username
>> datapath = '/mmfs1/home/<username>/Documents/MATLAB/Data';
>> % submit job
>> j = batch(c, 'preload_test', 'pool', 64, 'AdditionalPaths', datapath);
additionalSubmitArgs =
'--ntasks=65 --cpus-per-task=1 --ntasks-per-core=1'
>> x = j.fetchOutputs;
>> % look at serial execution time
>> x{1,1}.t1
ans =
91.2997
>> % look at parallel execution time
>> x{1,1}.t2
ans =
6.9289
>> % delete job
>> j.delete
Debugging
If a serial job produces an error, call the getDebugLog method to view the error log file. When submitting independent jobs, with multiple tasks, specify the task number.
>> c.getDebugLog(job.Tasks(3))
For Pool jobs, only specify the job object.
>> c.getDebugLog(job)
When troubleshooting a job, the cluster admin may request the scheduler ID of the job. This can be derived by calling schedID.
>> schedID(job)
ans =
25539
To Learn More
To learn more about the MATLAB Parallel Computing Toolbox, check out these resources:


